Yersinia pestis
| Yersinia pestis | |
|---|---|
 |
|
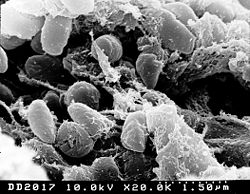
| A scanning electron microscope micrograph depicting a mass of Yersinia pestis bacteria. | |
| Scientific classification | |
| Domain: | Bacteria |
| Kingdom: | Eubacteria |
| Phylum: | Proteobacteria |
| Class: | Gammaproteobacteria |
| Order: | Enterobacteriales |
| Family: | Enterobacteriaceae |
| Genus: | Yersinia |
| Species: | Y. pestis |
| Binomial name | |
| Yersinia pestis (Lehmann & Neumann, 1896) van Loghem 1944 |
|
Yersinia pestis (formerly Pasteurella pestis) is a Gram-negative rod-shaped bacterium belonging to the family Enterobacteriaceae. It is a facultative anaerobe that can infect humans and other animals.[1]
Human Y. pestis infection takes three main forms: pneumonic, septicemic, and the notorious bubonic plagues.[1] All three forms are widely believed to have been responsible for a number of high-mortality epidemics throughout human history, including the Plague of Justinian in 542 and the Black Death that accounted for the death of at least one-third of the European population between 1347 and 1353. More recently, Y. pestis has gained attention as a possible biological warfare agent and the CDC has classified it as a category A pathogen requiring preparation for a possible terrorist attack.
Y. pestis was discovered in 1894 by Alexandre Yersin, a Swiss/French physician and bacteriologist from the Pasteur Institute, during an epidemic of plague in Hong Kong.[2] Yersin was a member of the Pasteur school of thought. Shibasaburo Kitasato, a German-trained Japanese bacteriologist who practiced Koch's methodology was also engaged at the time in finding the causative agent of plague.[3] However, it was Yersin who actually linked plague with Yersinia pestis. Originally named Pasteurella pestis, the organism was renamed in 1967.
Originally three biovars of Y. pestis were thought to correspond to one of the historical pandemics of bubonic plague.[4] Biovar Antiqua is thought to correspond to the Plague of Justinian; it is not known whether this biovar also corresponds to earlier, smaller epidemics of bubonic plague, or whether these were even truly bubonic plague.[5] Biovar Mediaevalis is thought to correspond to the Black Death. Biovar Orientalis is thought to correspond to the Third Pandemic and the majority of modern outbreaks of plague. However, calculations of Y pestis's evolutionary age, found using the number of synonymous single nucleotide polymorphisms (SNPs) in conjunction with molecular clock rates, date the emergence of the biovars prior to any of the historical epidemics due to the length of time needed to accumulate such mutations .[6] Additional evidence against this hypothesis includes the fact that Medievalis is likely too young to have produced the Black Death due to its recent divergence from Orientalis.[7]
Every year, thousands of cases of plague are still reported to the World Health Organization although with proper treatment, the prognosis for victims is now much better. A five to sixfold increase in cases occurred in Asia during the time of the Vietnam war, possibly due to the disruption of ecosystems and closer proximity between people and animals. Plague also has a detrimental effect on non-human mammals. In the United States of America, endangered animals such as the black-tailed prairie dog and the black-footed ferret are both under threat from the disease.
Contents |
Role in Black Death
Confirmed presence of Y. pestis would suggest that it was a contributing factor in some of (though possibly not all) the European plagues.
In 2000, Didier Raoult and others reported finding Y. pestis DNA by performing a "suicide PCR" on tooth pulp tissue from a fourteenth-century plague cemetery in Montpellier.[8]
However, in 2009 geneticists at Oxford University argued Raoult's approach was inadequate and reported having been unable to obtain any Y. pestis DNA from 121 teeth from sixty-six skeletons found in fourteenth-century mass graves. Lead author Alan Cooper concluded that though "[w]e cannot rule out Yersinia as the cause of the Black Death ...right now there is no molecular evidence for it." [9] [10]
General characteristics
Y. pestis is a rod-shaped facultative anaerobe with bipolar staining (giving it a safety pin appearance).[11] Similar to other Yersinia members, it tests negative for urease, lactose fermentation, and indole.[12] The closest relative is the gastrointestinal pathogen Yersinia pseudotuberculosis, and more distantly Yersinia enterocolitica.
Genome
The complete genomic sequence is available for two of the three sub-species of Y. pestis: strain KIM (of biovar Medievalis),[13] and strain CO92 (of biovar Orientalis, obtained from a clinical isolate in the United States).[14] As of 2006, the genomic sequence of a strain of biovar Antiqua has been recently completed.[15] Similar to the other pathogenic strains, there are signs of loss of function mutations. The chromosome of strain KIM is 4,600,755 base pairs long; the chromosome of strain CO92 is 4,653,728 base pairs long. Like its cousins Y. pseudotuberculosis and Y. enterocolitica, Y. pestis is host to the plasmid pCD1. In addition, it also hosts two other plasmids, pPCP1 (also called pPla or pPst) and pMT1 (also called pFra) which are not carried by the other Yersinia species. pFra codes for a phospholipase D that is important for the ability of Y. pestis to be transmitted by fleas.[16] pPla codes for a protease, Pla, that activates plasminogen in human hosts and is a very important virulence factor for pneumonic plague.[17] Together, these plasmids, and a pathogenicity island called HPI, encode several proteins which cause the pathogenesis, for which Y. pestis is famous. Among other things, these virulence factors are required for bacterial adhesion and injection of proteins into the host cell, invasion of bacteria in the host cell (via a Type III Secretion System), and acquisition and binding of iron that is harvested from red blood cells (via siderophores). Y. pestis is thought to be descendant from Y. pseudotuberculosis, differing only in the presence of specific virulence plasmids.
A comprehensive and comparative proteomics analysis of Y. pestis: strain KIM was performed in 2006.[18] The analysis focused on the transition to a growth condition mimicking growth in host cells.
Pathogenics and immunity
In the urban and sylvatic (forest) cycles of Y. pestis, most of the spreading occurs between rodents and fleas. In the sylvatic cycle, the rodent is wild, but in the urban cycle, the rodent is domestic. Additionally Y. pestis can spread from the urban environment and back again. Every infected animal can transmit the infection to humans through contact with skin tissue. Humans can also spread the bacteria to other humans through sneezing, coughing or direct contact with infected tissue.
In reservoir hosts
The reservoir commonly associated with Y. pestis are several species of rodents. In the steppes, the reservoir species is principally believed to be the marmot. In the United States, several species of rodents are thought to maintain Y. pestis. However, the expected disease dynamics have not been found in any rodent species. It is known that rodent populations will have a variable resistance, which could lead to a carrier status in some individuals.[19] There is evidence that fleas from other mammals have a role in human plague outbreaks.[20]
This lack of knowledge of the dynamics of plague in mammal species is also true among susceptible rodents such as the black-tailed prairie dog (Cynomys ludovicianus), in which plague can cause colony collapse resulting in a massive effect on prairie food webs.[21] However, the transmission dynamics within prairie dogs does not follow the dynamics of blocked fleas; carcasses, unblocked fleas, or another vector could possibly be important instead.[22]
In other regions of the world the reservoir of the infection is not clearly identified, which complicates prevention and early warning programs. One such example was seen in a 2003 outbreak in Algeria.[23]
Infector
The transmission of Y. pestis by fleas is well characterized.[24] Initial acquisition of Y. pestis by the vector occurs during feeding on an infected animal. Several proteins then contribute to the maintenance of the bacteria in the flea digestive tract, among them the hemin storage (Hms) system and Yersinia murine toxin (Ymt).
Although Yersinia murine toxin is highly toxic to rodents and was once thought to be produced to ensure reinfection of new hosts, it has been demonstrated that Ymt is important for the survival of Y. pestis in fleas.[16]
The Hms system plays an important role in the transmission of Y. pestis back to a mammalian host.[25] While in the insect vector, proteins encoded by Hms genetic loci induce biofilm formation in the proventriculus, a valve connecting the midgut to the esophagus.[26] Aggregation in the biofilm inhibits feeding and causes the flea to regurgitate blood. Transmission of Y. pestis occurs during the futile attempts of the flea to feed. Ingested blood is pumped into the esophagus, where it dislodges bacteria growing there and is regurgitated back into the host circulatory system.
In humans and other susceptible hosts
Pathogenesis due to Y. pestis infection of mammalian hosts is due to several factors including an ability of these bacteria to suppress and avoid normal immune system responses such as phagocytosis and antibody production. Flea bites allow for the bacteria to pass the skin barrier. Y. pestis expresses the yadBC gene, which is similar to adhesins in other Yersinia species, allowing for adherence and invasion of epithelial cells.[27] Y. pestis expresses a plasminogen activator that is an important virulence factor for pneumonic plague and which might degrade on blood clots in order to facilitate systematic invasion.[17] Many of the bacteria's virulence factors are anti-phagocytic in nature. Two important anti-phagocytic antigens, named F1 (Fraction 1) and V or LcrV, are both important for virulence.[11] These antigens are produced by the bacterium at normal human body temperature. Furthermore, Y. pestis survives and produces F1 and V antigens while it is residing within white blood cells such as monocytes, but not in neutrophils. Natural or induced immunity is achieved by the production of specific opsonic antibodies against F1 and V antigens; antibodies against F1 and V induce phagocytosis by neutrophils.[28]
Additionally, the Type III secretion system (T3SS) allows Y. pestis to inject proteins into macrophages and other immune cells. These T3SS-injected proteins are called Yops (Yersinia Outer Proteins) and include Yop B/D which form pores in the host cell membrane and have been linked to cytolysis. The YopO, YopH, YopM, YopT, YopJ and YopE are injected into the cytoplasm of host cells via T3SS into the pore created in part by YopB and YopD.[29] The injected Yop proteins limit phagocytosis and cell signaling pathways important in the innate immune system, as discussed below. In addition, some Y. pestis strains are capable of interfering with immune signaling (e.g. by preventing the release of some cytokines).
Yersinia pestis proliferates inside lymph nodes where it is able to avoid destruction by cells of the immune system such as macrophages. The ability of Yersinia pestis to inhibit phagocytosis allows it to grow in lymph nodes and cause lymphadenopathy. YopH is a protein tyrosine phosphatase that contributes to the ability of Yersinia pestis to evade immune system cells.[30] In macrophages, YopH has been shown to dephosphorylate p130Cas, Fyb (Fyn binding protein) SKAP-HOM and Pyk, a tyrosine kinase homologous to FAK. YopH also binds the p85 subunit of phosphoinositide 3-kinase, the Gab1 and Gab2 adapter proteins, and the Vav guanine nucleotide exchange factor.
YopE functions as a GTPase activating protein for members of the Rho family of GTPases such as RAC1. YopT is a cysteine protease that inhibits RhoA by removing the isoprenyl group which is important for localizing the protein to the cell membrane. It has been proposed that YopE and YopT may function to limit YopB/D-induced cytolysis.[31] This might limit the function of YopB/D to create the pores used for Yop insertion into host cells and prevent YopB/D-induced rupture of host cells and release of cell contents that would attract and stimulate immune system responses.
YopJ is an acetyltransferase that binds to a conserved α-helix of MAPK kinases.[32] YopJ acetylates MAPK kinases at serines and threonines that are normally phosphorylated during activation of the MAP kinase cascade.[33] [34] YopJ is activated in eukaryotic cells by interaction with target cell Phytic acid (IP6).[35] This disruption of host cell protein kinase activity causes apoptosis of macrophages, and it has been proposed that this is important for the establishment of infection and for evasion of the host immune response. YopO is a protein kinase also known as Yersinia protein kinase A (YpkA). YopO is a potent inducer of human macrophage apoptosis.[36]
Immunity
A formalin-inactivated vaccine once was available for adults at high risk of contracting the plague until removal from the market by the U.S. Food and Drug Administration. It was of limited effectiveness and may cause severe inflammation. Experiments with genetic engineering of a vaccine based on F1 and V antigens are underway and show promise. However, bacteria lacking antigen F1 are still virulent, and the V antigens are sufficiently variable, that vaccines composed of these antigens may not be fully protective.[37] United States Army Medical Research Institute of Infectious Diseases (USAMRIID) have found that an experimental F1/V antigen based vaccine protect cynomolgus macaques, but fails to protect African green monkeys.[38]
Clinical aspects
Symptoms and disease progression
- Bubonic plague
- Incubation period of 2–6 days, when the bacteria is actively replicating.
- Universally a general lack of energy
- Fever
- Headache and chills occur suddenly at the end of the incubation period
- Swelling of lymph nodes resulting in buboes, the classic sign of bubonic plague
- Septicemic plague
- Hypotension
- Hepatosplenomegaly
- Delirium
- Seizures in children
- Shock
- Universally a general lack of energy
- Fever
- Symptoms of bubonic or pneumonic plague are not always present
- Pneumonic plague
- Fever
- Chills
- Cough
- Chest pain
- Dyspnea
- Hemoptysis
- Lethargy
- Hypotension
- Shock
- Symptoms of bubonic or septicemic plague are not always present[39]
If this occurs with the classic buboes, this is considered primary, while secondary occurs after symptoms of bubonic or pneumonic infection. Since the bacteria are blood-borne, several organs can be affected including the spleen and brain. The diffuse infection can cause an immunologic cascade to occur, leading to disseminated intravascular coagulation (DIC), which in turn results in bleeding and necrotic skin and tissue. Such a disseminated infection increases mortality to 22%.
With the exception of the buboes, the initial symptoms of plague are very similar to many other diseases, making diagnosis difficult.[40]
ICD-9 codes for the diseases caused by Y. pestis:
- 020.0 Bubonic plague
- 020.2 Septicemic plague
- 020.5 Unspecified pneumonic plague
- 020.3 Primary pneumonic plague
- 020.4 Secondary pneumonic plague
Clinical determination
Grams stains can confirm the presence of gram negative rods, and in some cases the identification of the double curved shape. An anti-F1 serology test, which can differentiate between different species of Yersinia. Polymerase chain reaction (PCR) can be used to identify Y. pestis.
Treatment
The traditional first line treatment for Y. pestis has been streptomycin,[41][42] chloramphenicol, tetracycline,[43] and fluoroquinolones. There is also good evidence to support the use of doxycycline or gentamicin.[44] Resistant strains have been isolated; treatment should be guided by antibiotic sensitivities where available. Antibiotic treatment alone is insufficient for some patients, who may also require circulatory, ventilator, or renal support.
In an emergency department setting, Harrison's Principles of Internal Medicine outlines the following treatment course.[45] Antibiotics within the first 24 hours are very beneficial, with intravenous being preferred in pulmonary or advanced cases. Streptomycin or gentamicin are the first-line drugs, with chloramphenicol for critically ill patients, or rarely for suspected neuro-involvement.
Recent events
In September 2009, the death of Malcolm Casadaban, a molecular genetics professor at the University of Chicago, was linked to his work on a weakened laboratory strain of Y. pestis.[46]
Notes
- ↑ 1.0 1.1 Ryan KJ, Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. pp. 484–488. ISBN 0-8385852-9-9.
- ↑ Bockemühl J (1994). "[100 years after the discovery of the plague-causing agent--importance and veneration of Alexandre Yersin in Vietnam today]". Immun Infekt 22 (2): 72–5. PMID 7959865.
- ↑ Howard-Jones N (1973). "Was Shibasaburo Kitasato the discoverer of the plague bacillus?". Perspect Biol Med 16 (2): 292–307. PMID 4570035.
- ↑ Zhou D, Tong Z, Song Y, Han Y, Pei D, Pang X, Zhai J, Li M, Cui B, Qi Z, Jin L, Dai R, Du Z, Wang J, Guo Z, Wang J, Huang P, Yang R (2004). "Genetics of metabolic variations between Yersinia pestis biovars and the proposal of a new biovar, microtus". J Bacteriol 186 (15): 5147–52. doi:10.1128/JB.186.15.5147-5152.2004. PMID 15262951. PMC 451627. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=8195371.
- ↑ Guiyoule A, Grimont F, Iteman I, Grimont P, Lefèvre M, Carniel E (1994). "Plague pandemics investigated by ribotyping of Yersinia pestis strains". J Clin Microbiol 32 (3): 634–41. PMID 8195371.
- ↑ Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia et al. (2001). "Genome sequence of Yersinia pestis, the causative agent of plague". Nature 413 (6855): 523–527. doi:10.1038/35097083. PMID 11586360.
- ↑ Achtman, M., G. Morelli, P. X. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler et al (2004). "Microevolution and history of the plague bacillus, Yersinia pestis". Proceedings of the National Academy of Sciences of the United States of America 96: 17837–17842.
- ↑ Drancourt M, Aboudharam G, Signolidagger M, Dutourdagger O, Raoult D. (2002). "Detection of 400-year-old Yersinia pestis DNA in human dental pulp: An ory of plague.". Microbes Infect. 4 (1): 105–9. doi:10.1016/S1286-4579(01)01515-5. PMID 11825781.
- ↑ (PDF) Absence of Y. pestis-specific DNA in human teeth from European excavations of putative plague victims. British Society for General Microbiology. https://www.sgm.ac.uk/meetings/pdfabstracts/umist2003abs.pdf.
- ↑ "Jury out on Black Death culprit". BBC News. 10 September 2003. http://news.bbc.co.uk/2/hi/health/3097934.stm. Retrieved 2008-12-12.
- ↑ 11.0 11.1 Collins FM (1996). Pasteurella, Yersinia, and Francisella. In: Baron's Medical Microbiology (Baron S et al, eds.) (4th ed.). Univ. of Texas Medical Branch. ISBN 0-9631172-1-1. http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mmed.section.1611.
- ↑ Stackebrandt, Erko; Dworkin, Martin; Falkow, Stanley; Rosenberg, Eugene; Karl-Heinz Schleifer (2005). The Prokaryotes: A Handbook on the Biology of Bacteria:Volume 6: Proteobacteria: Gamma Subclass. Berlin: Springer. ISBN 0-387-25499-4.
- ↑ Deng W et al.. (2002). "Genome Sequence of Yersinia pestis KIM". Journal of Bacteriology 184 (16): 4601–4611. doi:10.1128/JB.184.16.4601-4611.2002. PMID 12142430.
- ↑ Parkhill J et al.. (2001). "Genome sequence of Yersinia pestis, the causative agent of plague". Nature 413 (6855): 523–527. doi:10.1038/35097083. PMID 11586360.
- ↑ Chain PS, Hu P, Malfatti SA, et al (2006). "Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen". J. Bacteriol. 188 (12): 4453–63. doi:10.1128/JB.00124-06. PMID 16740952.
- ↑ 16.0 16.1 Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A (2002). "Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector". Science 296 (5568): 733–5. doi:10.1126/science.1069972. PMID 11976454.
- ↑ 17.0 17.1 Lathem WW, Price PA, Miller VL, Goldman WE (2007). "A plasminogen-activating protease specifically controls the development of primary pneumonic plague". Science 315 (5811): 509–13. doi:10.1126/science.1137195. PMID 17255510.
- ↑ Hixson K et al.. (2006). "Biomarker candidate identification in Yersinia pestis using organism-wide semiquantitative proteomics.". Journal of Proteome Research 5 (11): 3008–3017. doi:10.1021/pr060179y. PMID 16684765.
- ↑ MEYER KF (1957). "The natural history of plague and psittacosis". Public Health Rep 72 (8): 705–19. PMID 13453634.
- ↑ von Reyn CF, Weber NS, Tempest B, et al (1977). "Epidemiologic and clinical features of an outbreak of bubonic plague in New Mexico". J. Infect. Dis. 136 (4): 489–94. PMID 90884.
- ↑ Pauli JN, Buskirk SW, Williams ES, Edwards WH (2006). "A plague epizootic in the black-tailed prairie dog (Cynomys ludovicianus)". J. Wildl. Dis. 42 (1): 74–80. PMID 16699150.
- ↑ Webb CT, Brooks CP, Gage KL, Antolin MF (2006). "Classic flea-borne transmission does not drive plague epizootics in prairie dogs". Proc. Natl. Acad. Sci. U.S.A. 103 (16): 6236–41. doi:10.1073/pnas.0510090103. PMID 16603630.
- ↑ Bertherat E, Bekhoucha S, Chougrani S, et al (2007). "Plague Reappearance in Algeria after 50 Years, 2003". Emerging Infect. Dis. 13 (10): 1459–1462. PMID 18257987.
- ↑ Zhou D, Han Y, Yang R (2006). "Molecular and physiological insights into plague transmission, virulence and etiology". Microbes Infect. 8 (1): 273–84. doi:10.1016/j.micinf.2005.06.006. PMID 16182593.
- ↑ B.J. Hinnebusch, R.D. Perry and T.G. Schwan (1996). "Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas". Science. 8 (1): 367–70. doi:10.1126/science.273.5273.367. PMID 8662526.
- ↑ Erickson, D. L., N. R. Waterfield, V. Vadyvaloo, D. Long, E. R. Fischer, R. ffrench-Constant, and B. J. Hinnebusch (2007). "Acute oral toxicity of yersinia pseudotuberculosis to fleas: Implications for the evolution of vector-borne transmission of plague.". Cellular Microbiology 9 (11): 2658–2666. doi:10.1111/j.1462-5822.2007.00986.x. PMID 17587333.
- ↑ Forman S, Wulff CR, Myers-Morales T, Cowan C, Perry RD, Straley SC (2008). "yadBC of Yersinia pestis, a new virulence determinant for bubonic plague". Infect. Immun. 76 (2): 578–87. doi:10.1128/IAI.00219-07. PMID 18025093.
- ↑ Salyers AA, Whitt DD (2002). Bacterial Pathogenesis: A Molecular Approach (2nd ed.). ASM Press. pp. 207-12.
- ↑ Viboud GI, Bliska JB (2005). "Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis". Annu. Rev. Microbiol. 59: 69–89. doi:10.1146/annurev.micro.59.030804.121320. PMID 15847602.
- ↑ de la Puerta ML, Trinidad AG, del Carmen Rodríguez M, Bogetz J, Sánchez Crespo M, Mustelin T, Alonso A, Bayón Y (February 2009). "Characterization of new substrates targeted by Yersinia tyrosine phosphatase YopH". PLoS ONE 4 (2): e4431. doi:10.1371/journal.pone.0004431. PMID 19221593. PMC 2637541. http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0004431.
- ↑ Mejía E, Bliska JB, Viboud GI (February 2009). "Yersinia controls type III effector delivery into host cells by modulating Rho activity". PLoS ONE 4 (2): e4431. doi:10.1371/journal.ppat.0040003. PMID 18193942. PMC 2186360. http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0004431.
- ↑ Hao YH, Wang Y, Burdette D, Mukherjee S, Keitany G, Goldsmith E, Orth K (January 2008). "Structural requirements for Yersinia YopJ inhibition of MAP kinase pathways". PLoS ONE 2 (3): e1375. doi:10.1371/journal.pone.0001375. PMID 18167536. PMC 2147050. http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0001375.
- ↑ Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K (May 2006). "Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation". Science 312 (5777): 1211–1214. doi:10.1126/science.1126867. PMID 16728640. http://www.sciencemag.org/cgi/content/full/312/5777/1211.
- ↑ Mittal R, Peak-Chew S-Y, McMahon HT (December 2006). "Acetylation of MEK2 and IκB kinase (IKK) activation loop residues by YopJ inhibits signalling". Proc Natl Acad Sci 103 (49): 18574–18579. doi:10.1073/pnas.0608995103. PMID 17116858. PMC 1654131. http://www.pnas.org/content/103/49/18574.long.
- ↑ Mittal R, Peak-Chew SY, Sade RS, Vallis Y, McMahon HT (2010). "The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate". J Biol Chem 285 (26): 19927–34. doi:10.1074/jbc.M110.126581. PMID 20430892. PMC 2888404. http://www.jbc.org/content/early/2010/04/29/jbc.M110.126581.long.
- ↑ Park H, Teja K, O'Shea JJ, Siegel RM (May 2007). "The Yersinia effector protein YpkA induces apoptosis independently of actin depolymerization". J Immunol. 178 (10): 6426–6434. PMID 17475872. http://www.jimmunol.org/cgi/content/full/178/10/6426.
- ↑ Welkos S et al.. (2002). "Determination of the virulence of the pigmentation-deficient and pigmentation-/plasminogen activator-deficient strains of Yersinia pestis in non-human primate and mouse models of pneumonic plague". Vaccine 20 (17-18): 2206–2214. doi:10.1016/S0264-410X(02)00119-6. PMID 12009274.
- ↑ Pitt ML (October 13–14). Non-human primates as a model for pneumonic plague. In: Animals Models and Correlates of Protection for Plague Vaccines Workshop.
- ↑ Info taken from "Harrison's Principles of Internal Medicine 16th Edition"
- ↑ Prentice MB, Rahalison L (2007). "Plague". Lancet 369 (9568): 1196–207. doi:10.1016/S0140-6736(07)60566-2. PMID 17416264. http://linkinghub.elsevier.com/retrieve/pii/S0140-6736(07)60566-2.
- ↑ Wagle PM. (1948). "Recent advances in the treatment of bubonic plague". Indian J Med Sci 2: 489–94.
- ↑ Meyer KF. (1950). "Modern therapy of plague". JAMA 144 (12): 982–5. PMID 14774219.
- ↑ Kilonzo BS, Makundi RH, Mbise TJ. (1992). "A decade of plague epidemiology and control in the Western Usambara mountains, north-east Tanzania". Acta Tropica 50 (4): 323–9. doi:10.1016/0001-706X(92)90067-8. PMID 1356303.
- ↑ Mwengee W, Butler T, Mgema S, et al. (2006). "Treatment of plague with gentamicin or doxycycline in a randomized clinical trial in Tanzania". Clin Infect Dis 42 (5): 614–21. doi:10.1086/500137. PMID 16447105.
- ↑ Jameson, J. N. St C.; Dennis L. Kasper; Harrison, Tinsley Randolph; Braunwald, Eugene; Fauci, Anthony S.; Hauser, Stephen L; Longo, Dan L. (2005). Harrison's principles of internal medicine. New York: McGraw-Hill Medical Publishing Division. ISBN 0-07-140235-7.
- ↑ Sadovi, Carlos (2009-09-19). "U. of C. researcher dies after exposure to plague bacteria". Chicago Breaking News Center. http://www.chicagobreakingnews.com/2009/09/uofc-researcher-dies-after-exposer-to-plague-bacteria.html. Retrieved 2010-03-03.
External links
- Yersinia pestis. Virtual Museum of Bacteria.
- A list of variant strains and information on synonyms (and much more) is available through the NCBI taxonomy browser.
- CDC's Home page for Plague [1]
- IDSA's resource page on Plague: Current, comprehensive information on pathogenesis, microbiology, epidemiology, diagnosis, and treatment[2]
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||